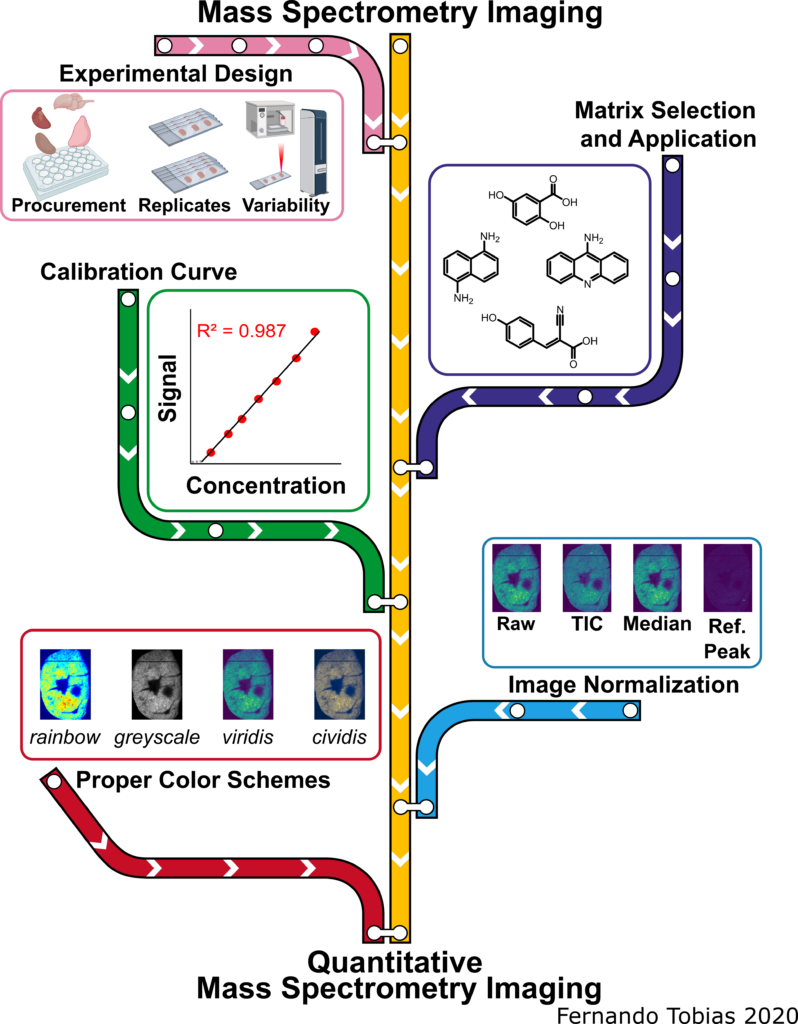
Welcome to the Hummon Research Group at the Ohio State University. Our research interests lie at the intersection of analytical chemistry and chemical biology, with a focus on cancer biology.
Current Projects:
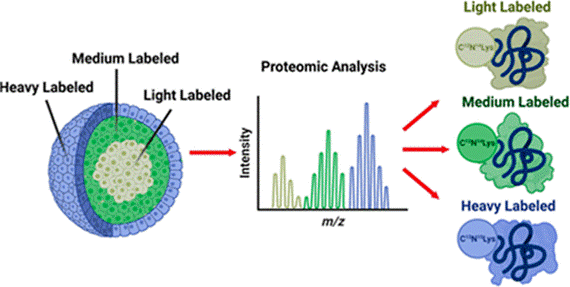
Spatial SILAC in Cellular Layers of the Spheroid
Stable isotopic labeling of amino acids in cell culture (SILAC) is a metabolic labeling method that identifies the origin of a protein in multiplexed experiments. By pulsing different heavy amino acids into cell culture media at set time points during spheroid growth, we can discretely label the cellular layers that form in these tumor models. Proteins from the necrotic core, quiescent middle layer, and proliferative outer layer of spheroids can be distinguished and quantified in a proteomic analysis, generating valuable information about chemotherapeutics’ effect on different cellular populations.
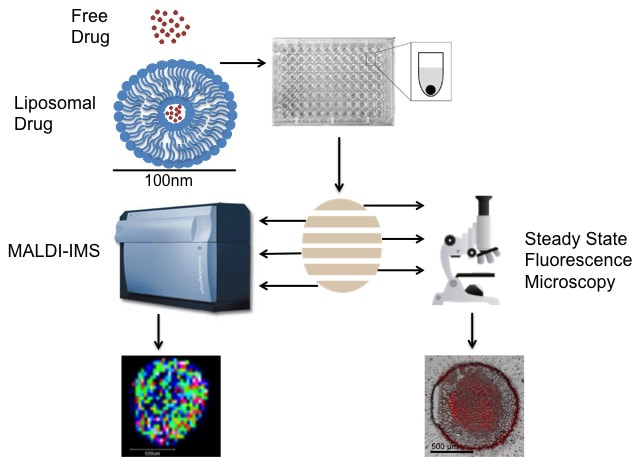
Analysis of Liposomal Drug Delivery Systems
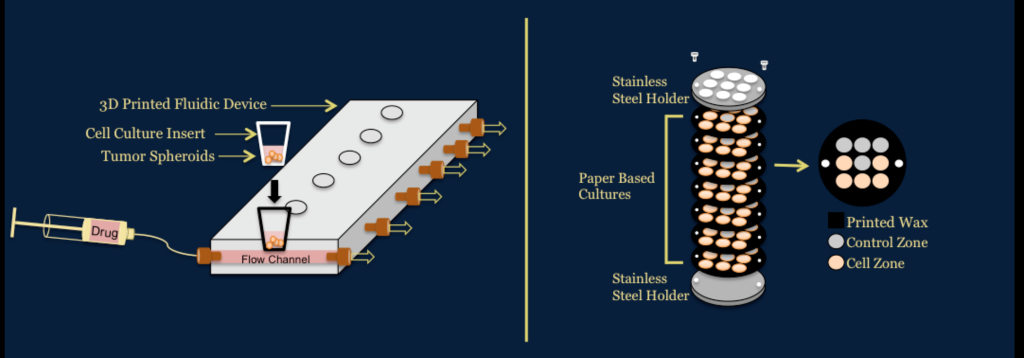
Investigation of Novel in vitro Platforms to Analyze the Efficacy and Toxicity of Chemotherapeutics
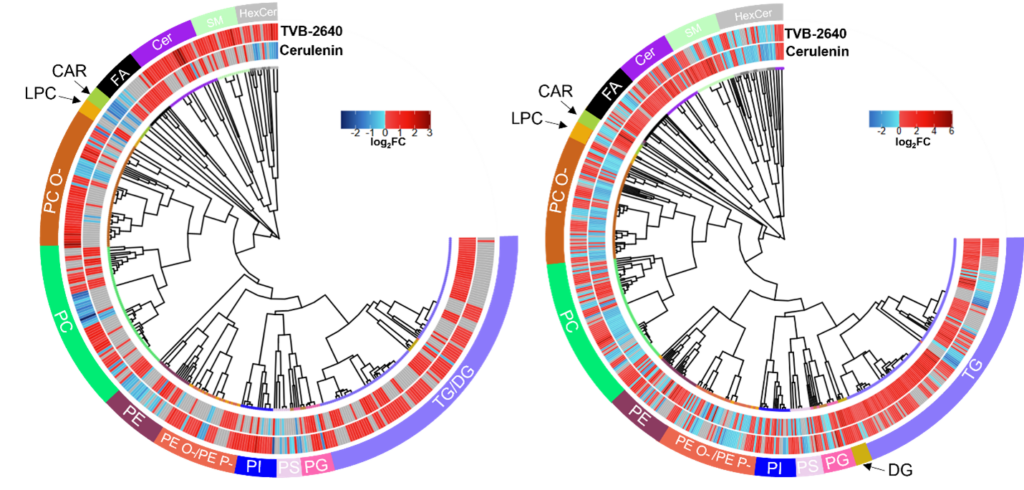
Lipidomics and proteomics of FASN inhibited colorectal cancer spheroids
FASN is often upregulated in a multitude of cancers and has become a druggable target of interest for the treatment of a multitude of cancers. However, the distinct molecular underpinnings of inhibiting this and other metabolic enzymes are unknown. Using both lipidomics and proteomics, we are able to determine the distinct molecular differences of different FAS inhibitors in two different sets of colorectal cancer spheroids and determine that each FAS inhibitor may be causing distinct death mechanisms to occur.