My group uses a wide variety of chemical and biophysical approaches to answer fundamental questions focusing on nucleic acids (RNA and DNA) and proteins that are involved in the translation of the genetic code and viral replication. Here, we describe the current interests of our Virus subgroup members.
1.Host Cellular Factors Critical for HIV-1 Replication. The primer for reverse transcription in HIV-1, human tRNALys3, is selectively packaged into virions along with tRNALys1,2. Human lysyl-tRNA synthetase (LysRS), the only cellular factor that interacts specifically with both tRNALys isoacceptors, is also packaged into HIV-1. Selective packaging of tRNALys depends on the ability of LysRS to bind to tRNALys and the presence of both host cell factors is required for optimal viral infectivity. LysRS is normally part of a dynamic mammalian multi-synthetase complex (MSC). We have shown that HIV-1 infection results in a free pool of phosphorylated LysRS (pS207-LysRS) that is partially re-localized to the nucleus of target cells. Blocking this pathway in HIV-1 producing cells abolished LysRS packaging and resulted in less infectious progeny virions. We are currently focusing on elucidating the various role of human LysRS in the HIV-1 lifecycle and determining the cellular location and conformational effects of tRNA primer annealing to genomic RNA.
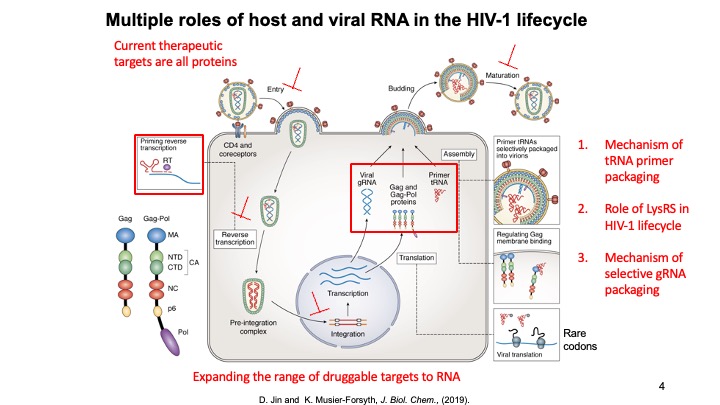
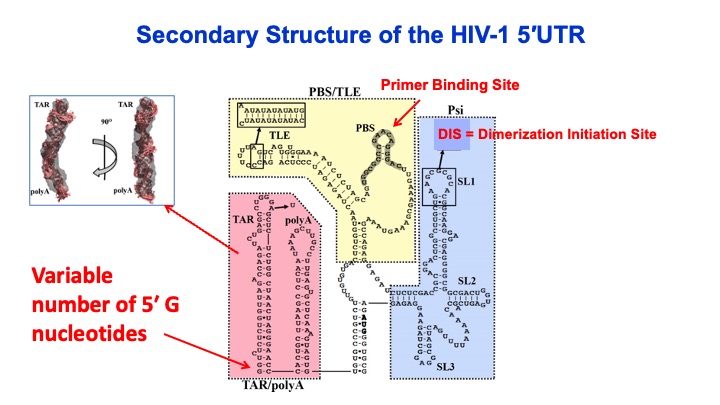
2. RNA Binding and Packaging by Retroviral Gag Proteins. HIV-1 packages two copies of full-length genomic RNA (gRNA) as a dimer into newly formed viral particles. The full-length RNA also serves as an mRNA for Gag polyprotein synthesis. ONe focus of this project is to understand how the fate of the viral RNA (gRNA vs. mRNA) is determined. Retroviral assembly doesn’t depend on the presence of gRNA, yet gRNA packaging is essential for production of infectious virus particles. Robust and specific mechanisms must therefore be in place to ensure the genetic material is passed on to progeny virions. While there are numerous therapeutics in clinical use, there are currently no gRNA packaging or viral assembly inhibitors. Selective gRNA packaging is facilitated by interactions between the HIV-1 Gag protein and conserved gRNA elements within the 5′ untranslated region (5′UTR). Recently, the variable number of G residues (1G, 2G and 3G) at the 5′ end of the HIV-1 RNA transcript has been reported to affect the gRNA localization, with 1G RNA preferentially selected over 3G to be the viral genome. This is very surprising given that these two 9-kb RNAs only differ by 2 nucleotides. We are focused on elucidating the mechanism by which transcriptional start site choice modulates gRNA packaging selectivity. This work will gain insights into how subtle sequence changes can alter the ensemble of 5′UTR RNA structures, HIV-1 Gag binding, and viral RNA packaging versus translation.
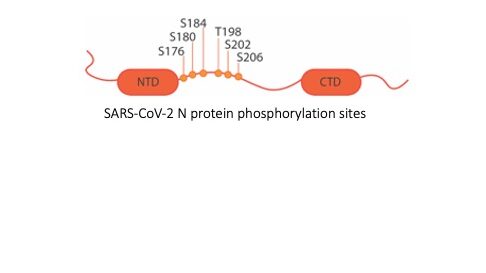
3. Role of SARS-CoV-2 N protein phosphorylation in RNA binding, condensation, and chaperone activity. SARS-CoV-2 is a betacoronavirus with a positive sense, single-stranded RNA genome about 30 kilobases (kb) in length. The viral genomic RNA (gRNA) is packaged into progeny virions by Nucleocapsid (N) protein, which distinguishes viral gRNA from other cellular RNAs by recognizing the genomic packaging signal (PS), which is not yet clearly defined. In addition, gaps remain in our understanding of the packaging mechanism. Since genome replication and packaging are both critical for viral infection to persist, investigating the molecular interactions specific to these processes will provide potential antiviral therapeutic targets. In this project, we are studying the intermolecular interactions between specific gRNA regions and SARS-CoV-2 N protein. In addition to its role in gRNA packaging, N protein is known to be an RNA chaperone during gRNA replication in other coronaviruses and post-translational phosphorylation of N protein is hypothesized to modulate its function. These hypotheses are also being tested in this work.