My group uses a wide variety of chemical and biophysical approaches to answer fundamental questions focusing on nucleic acids (RNA and DNA) and proteins that are involved in translation of the genetic code and viral replication. Here, we describe the current interests of our synthetase subgroup members.
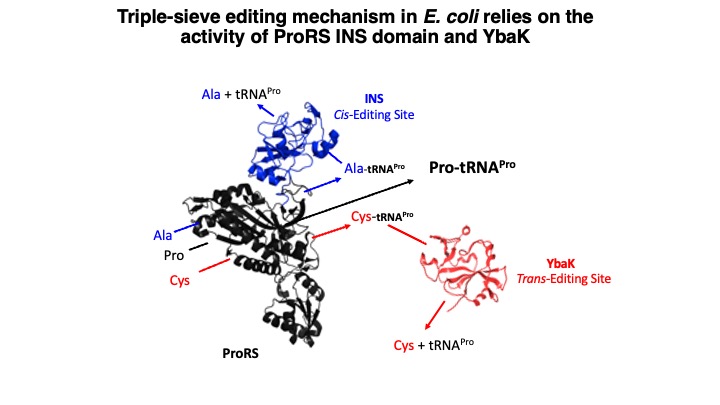
1.Translational Quality Control by Aminoacyl-tRNA synthetases. Aminoacyl-tRNA synthetases (aaRSs) are universally conserved enzymes that ensure high fidelity of translation of genetic information into functional proteins across all domains of life. These enzymes pair amino acids with their corresponding tRNA isoacceptors in a process known as aminoacylation. The size and functional group similarity among related amino acids make it challenging for aaRSs to accurately distinguish these small substrates. Despite the challenges in amino acid selection, errors in translation only occur about every 1 in 10,000 codons; this suggests the existence of proofreading mechanisms to edit the majority of aa-tRNA mischarging events prior to translation at the ribosome. Indeed, many aaRSs have acquired editing mechanisms that prevent formation and/or accumulation of mispaired tRNAs. Shown in the figure below is the “triple-sieve” editing mechanism we initially discovered in E. coli. Most bacteria encode a ProRS with an editing domain known as the insertion (INS) domain, which hydrolyzes misacylated Ala-tRNAPro. However, the INS domain can’t hydrolyze Cys-tRNAPro. Instead, many bacteria encode a separate editing domain known as YbaK that carries out this function.
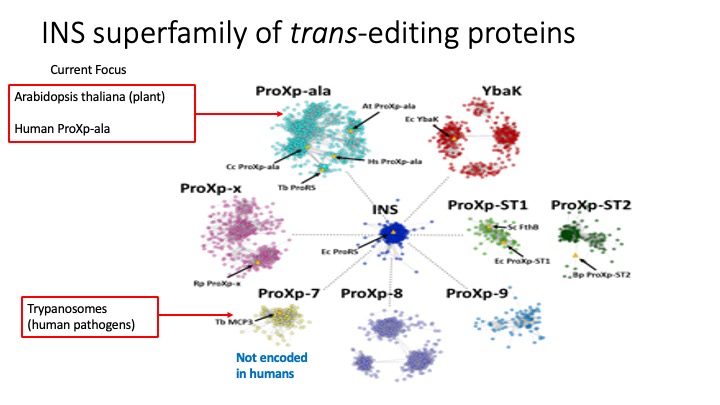
2. Translational Quality Control by Trans-editing Domains. Free-standing trans-editing domains that are structurally homologous to editing domains have been identified in all domains of life. The overarching goal of this project is to uncover the specific functions of a growing family of proteins known as the INS superfamily (see figure below). This diverse yet universally conserved family now has a solid and accumulating in vitro structure-function knowledge base, which strongly supports a role in maintaining translational fidelity. Our knowledge of the broader physiological roles of these proteins, especially in eukaryotes, is still in its infancy and is the focus of current work. The eukaryotic trans-editing systems we are currently investigating include plants, humans, and trypanosomes, which are human pathogens. Structure-function studies are being carried out both in vitro and using cell-based or in vivo approaches.
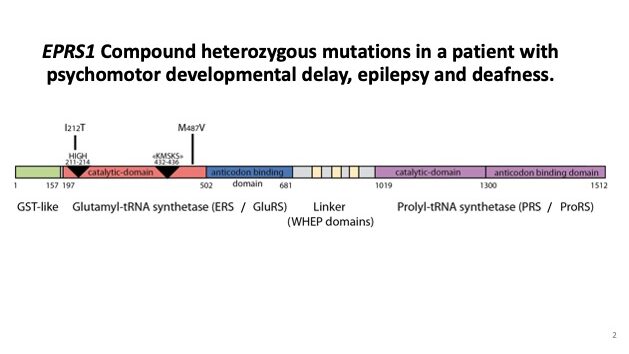
3. Role of Aminoacyl-tRNA Synthetases in Genetic Disease. Heritable mutations in genes encoding aminoacyl-tRNA synthetases are causally linked to certain diseases including cancer, neuronal pathologies, autoimmune disorders, and disrupted metabolic conditions. We are investigation variants of the dual-functional cytoplasmic human glutamyl-prolyl-tRNA synthetase, EPRS1, which have been identified by clinicians to be associated with leukodystrophy, diabetes and bone disease, psychomotor developmental delay, seizures and deafness. Our functional analyses focus on the impact of disease-associated mutations on enzymatic function, including tRNA binding and aminoacylation, as well as structural effects. Disruptions in protein homeostasis that arise in cells with tRNA charging defects can induce certain adaptive response pathways including the integrated stress response (ISR). We also determine whether there is altered induction of the ISR in cells expressing the mutant EPRS1.