Problem:
I am experiencing a strange problem with generating symmetry mates in PyMOL for the coordinate file 1WAP: specifically, the generated molecules clash with each other. I tried using either the menus Action > generate > symmetry mates …. , or using the symexp command; same result
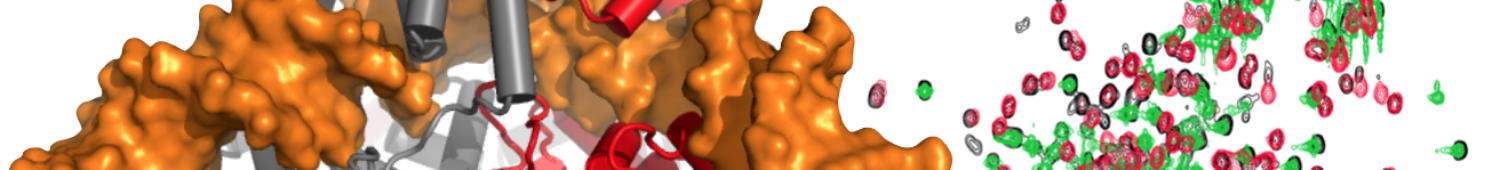
Coot is able to reconstruct the proper arrangements (see attached image).
Correspondence from the PyMOL Support team:
The behavior you are describing is due to PyMOL ignoring SCALE matrix translation. Unfortunately, this bug was accidentally introduced into recent versions of PyMOL. Our developers are aware of it and it will be fixed in the next release. Thank you for bringing it to our attention and apologies for the inconvenience.
In the meantime, you can use a work-around by explicitly applying ORIGX1 translation (the value is taken from the PDB header). This needs to be done before generating symmetry mates. I.e.:
fetch 1wap
as cartoon
alter_state 1, 1wap, x += 38.97750
symexp sym, 1wap, (1wap), 10
Give it a try and let us know if it doesn’t work for you or if you have additional questions and would like further assistance.
Best regards,
Pavel Golubkov
Correspondence from the Chimera Support team:
Yep Chimera does not get the correct crystal symmetry for 1WAP. The problem is Chimera is assuming that the fractional crystal unit cell coordinates origin is 0,0,0 in physical Angstrom coordinates. But the PDB format allows them to be different, and for 1WAP there is a shift by 0.25 along the x axis in fractional coordinates. This is seen in the SCALE1, SCALE2, SCALE3 records in the PDB format file and in the _atom_sites.fract_transf_vector table in the mmCIF format file. I didn’t know the PDB allowed this. A quick search of 900 random PDB files showed 2 with this problem: 1HHO from 1983 and 2DHB from 1972. 1WAP is from 1995. I guess this unfortunate coordinate system irregularity only effects very
old PDB entries.
Let me think about whether it is worth fixing this. We are putting all our effort into our next generation ChimeraX right now.
You can work around the bug by shifting the 1WAP coordinates 0.25 unit cells along x. Use menu Tools / Movement / Transform Coordinates, enter shift 38.9775 0 0, press Apply, then the Unit Cell tool and others that use space group symmetry will work correctly.
Goddard