Small molecules generally require well-defined, hydrophobic binding pockets in their target proteins to achieve high affinity and specificity (Figure 1a). This largely limits small molecules to targeting enzymes, G-protein coupled receptors, and ion channels. Protein-protein interactions (PPIs) usually involve large, flat binding sites, thus representing a challenging class of drug targets for conventional small molecules. This is because small molecules generally do not make enough points of contact with a flat surface (Figure 1b). On the other hand, we (and others) have previously demonstrated that macrocyclic peptides in the molecular-weight range of 500-2000 are capable of binding to flat surfaces with antibody-like affinity and specificity and serve as effective PPI inhibitors (Figure 1c). These macrocycles are 3-5 times larger than conventional small molecules and therefore make more pints of contact with a flat surface.
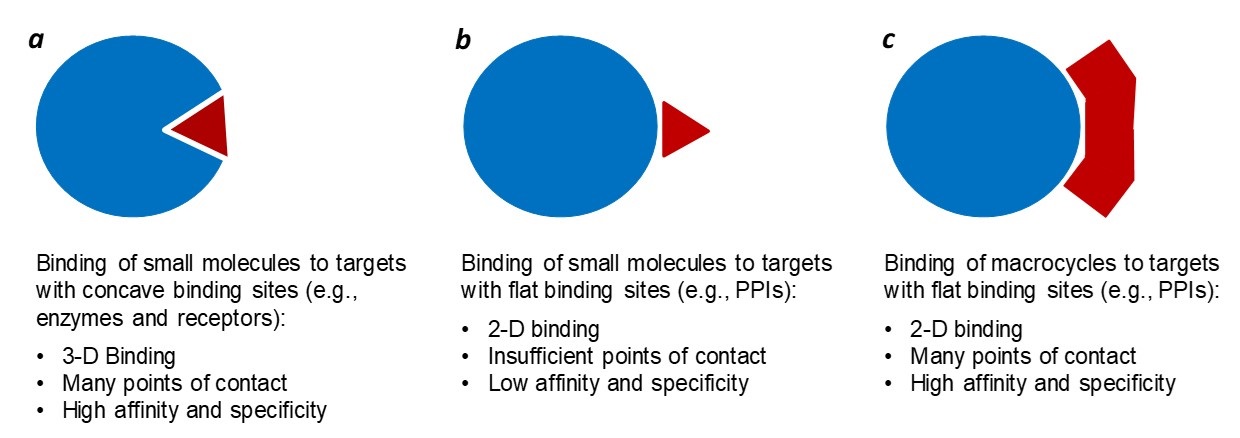
Figure 1. Comparison of small molecules and macrocycles for target binding. (a) A small-molecule inhibitor is engulfed by the deep binding pocket of a traditional target (e.g., an enzyme or receptor), resulting in high affinity and specificity. (b) Binding of a small molecule to a flat PPI binding site results in low affinity and specificity. (c) A macrocycle engages in many points of contact with a flat PPI binding site and achieves high affinity and specificity.
Unfortunately, macrocyclic ligands of the above size range are challenging to design by rational approaches. We therefore choose to chemically synthesize large combinatorial libraries of macrocyclic peptides 108 different compounds) and then rapidly screen them for binding to a protein of interest (Figure 2).
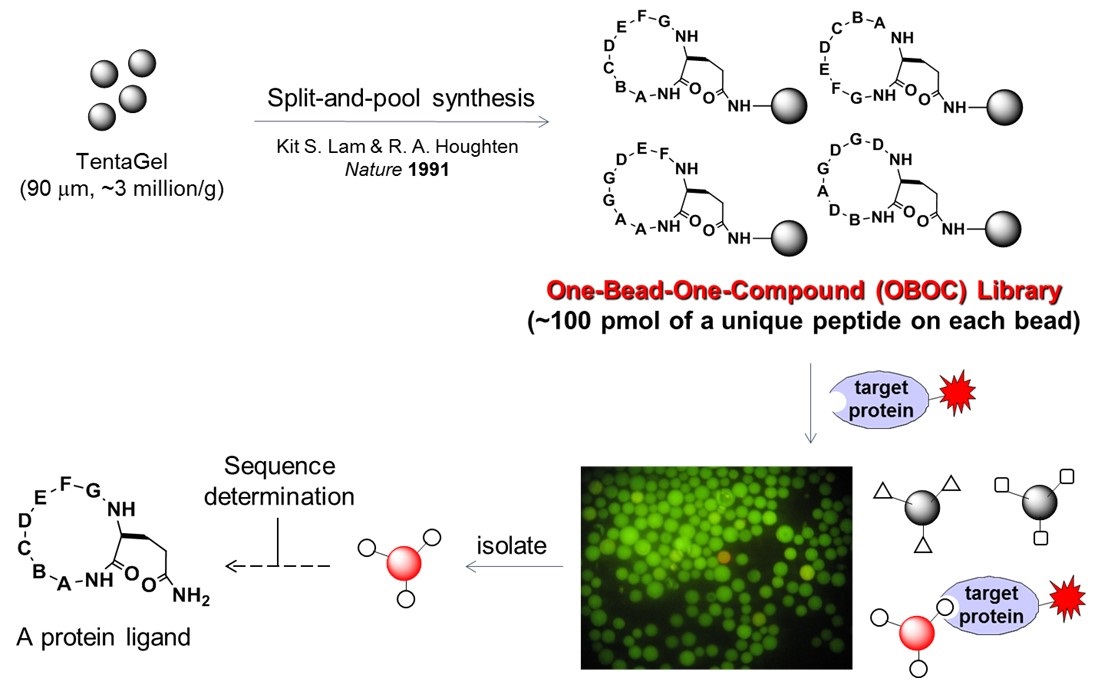
Figure 2. Scheme showing the combinatorial synthesis of cyclic peptides in the OBOC format and screening for binding against a fluorescently labeled protein.
We have developed novel methods to synthesize and screen peptide-encoded monocyclic [1] and bicyclic peptide libraries [2] on spatially segregated microbeads, in which each bead displays a unique cyclic (or bicyclic) peptide on its surface and a linear peptide of the same sequence as an encoding tag inside the bead (Figure 3).
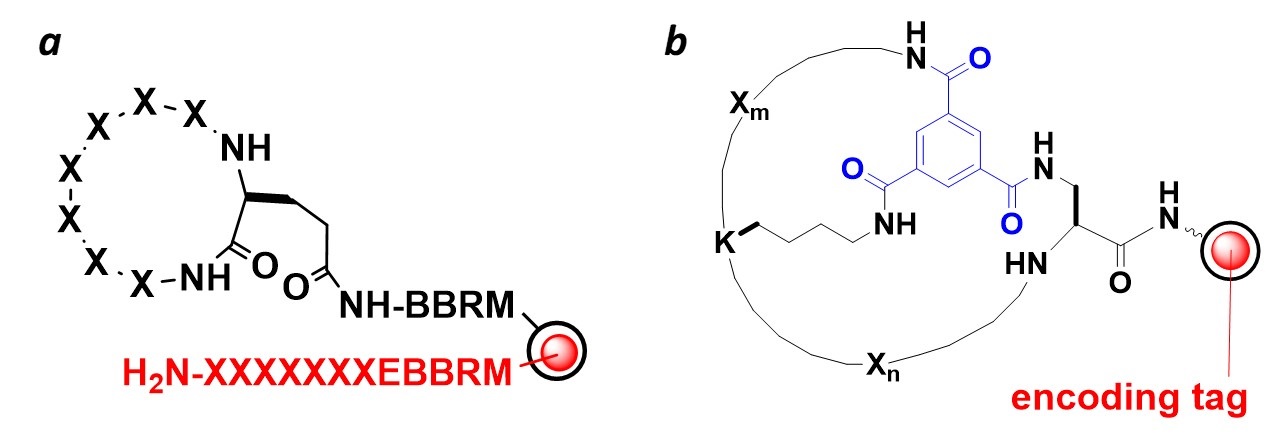
Figure 3. Macrocyclic peptide libraries in the one bead-two compound (OBTC) format, in which each bead displays a cyclic (a) or bicyclic peptide (b) on its surface and a linear peptide of identical sequence in its interior as an encoding tag.
After a positive hit is isolated, the structure of the macrocycle is determined by sequencing the encoding tag inside the bead by a partial Edman degradation-mass spectrometry (PED-MS) method [3]. Some examples of macrocyclic peptide ligands identified from our libraries are illustrated in Figure 4 [4-7].
Future research in this area includes:
1) Optimization of previous library hits (e.g., inhibitors against calcineurin, K-Ras, NEMO, -catenin, and PDZ domains) to improve their potency, selectivity, stability, and solubility
2) Screening libraries against additional disease-modifying proteins
Key Publications:
- Joo, S. H., Xiao, Q., Ling, Y., Gopishetty, B., and Pei, D. (2006) High-throughput sequence determination of cyclic peptide library members by partial Edman degradation/mass spectrometry. Am. Chem. Soc. 128, 13000-13009.
- Lian, W., Upadhyaya, P., Rhodes, C. A., Liu, Y., and Pei, D. (2013) Screening Bicyclic Peptide Libraries for Protein−Protein Interaction Inhibitors: Discovery of a Tumor Necrosis Factor-α J. Am. Chem. Soc. 135, 11990-11995.
- Thakkar, A., Wavreille, A.-S., and Pei, D. (2006) Traceless capping agent for peptide sequencing by partial Edman degradation and mass spectrometry. Chem. 78, 5935-5939.
- Liu, T.; Liu, Y.; Kao, H.-Y.; Pei, D. (2010) Membrane Permeable Cyclic Peptidyl Inhibitors against Human Peptidylprolyl Isomerase Pin1. Med. Chem. 53, 2494-2501.
- Liu, T., Qian, Z., Xiao, Q., and Pei, D. (2011) High-Throughput Screening of One-Bead-One-Compound Libraries: Identification of Cyclic Peptidyl Inhibitors against Calcineurin/NFAT Interaction. ACS Comb. Sci. 13, 537-546.
- Wu, X., Upadhyaya, P., Villalona-Calero, M. A., Briesewitz, R., and Pei, D. (2013) Inhibition of Ras-effector interactions by cyclic peptides. Chem. Commun. 4, 378-382.
- Upadhyaya, P., Qian, Z., Habir, N. A. A., and Pei, D. (2014) Direct Ras Inhibitors Identified from a Structurally Rigidified Bicyclic Peptide Library. Tetrahedron 70, 7714-7720.