Although most biomolecules (e.g., peptides, proteins, and nucleic acids) are impermeable to the cell membrane, viruses, bacterial protein toxins, and an increasing number of eukaryotic proteins are known to enter the cytosol of eukaryotic cells by crossing the plasma, endosomal, or ER membrane (Figure 1). In addition, some bacterial and eukaryotic proteins, after being synthesized inside the cytosol, are transported in their native states into the extracellular/periplasmic space or intracellular organelles (e.g., the peroxisome). How these biomolecules move across a cell membrane has been a longstanding mystery in biology. Solving this mystery is not only of great scientific importance but also have many industrial applications. For example, ~75% disease-relevant human proteins are out of the reach of current drug modalities, which include small molecules and biologics (e.g., monoclonal antibodies). Understanding how biomolecules travel across the cell membrane would help design cell-permeable biologics as a new class of drugs to target the ~75% undruggable proteins.
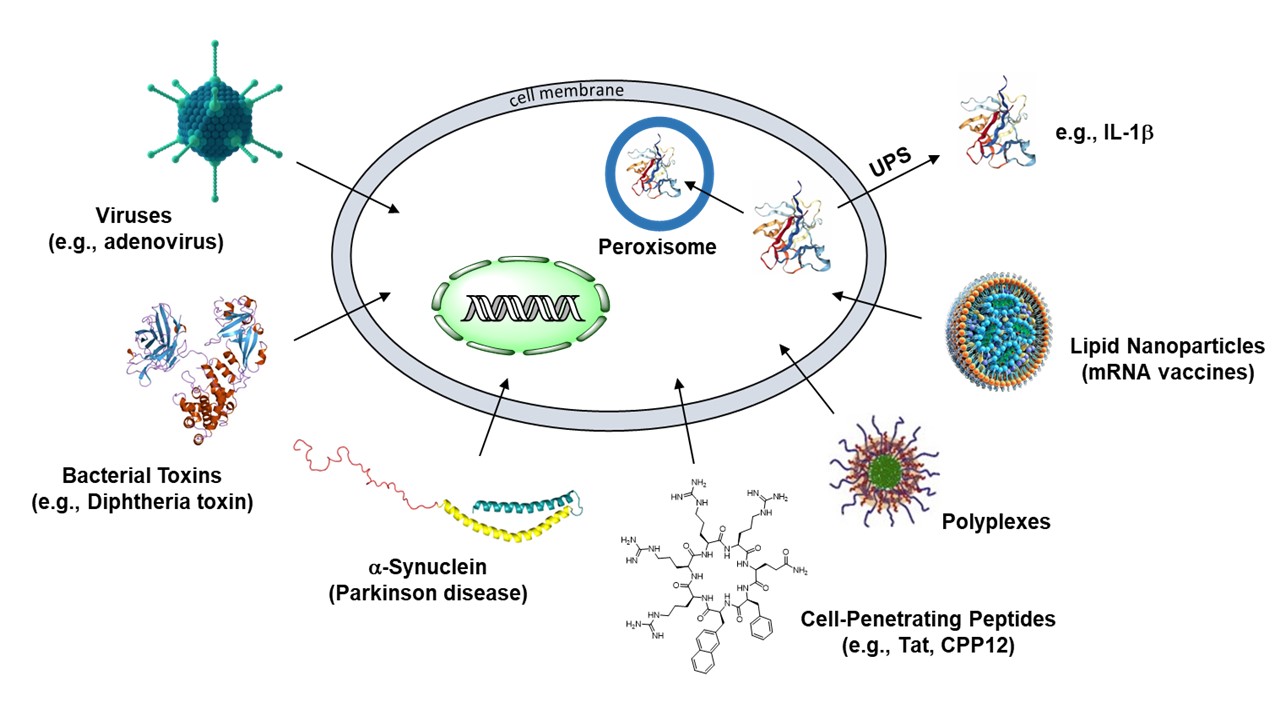
Figure 1. Different types of biomolecules/systems that are known to translocate across various cellular membranes. UPS, unconventional protein secretion.
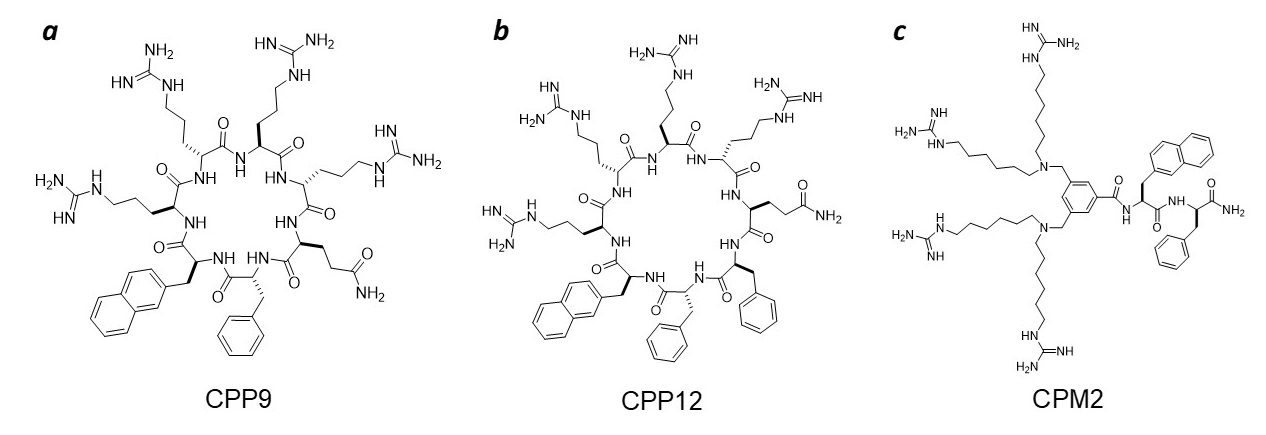
Our group is at the forefront of this important field. During the past decade, we discovered a family of small amphipathic cyclic peptides as exceptionally active cell-penetrating peptides (CPPs), e.g., cyclo(phe-Nal-Arg-arg-Arg-arg-Gln) (CPP9) and cyclo(Phe-phe-Nal-Arg-arg-Arg-arg-Gln) (CPP12) (Figure 2) [1]. These cyclic CPPs exhibit excellent cytosolic entry efficiencies (up to 120%), proteolytic stability, bioavailability, and broad biodistribution.
Our mechanistic studies revealed that CPPs bind directly to the plasma membrane phospholipids, enter the cell by endocytosis, and then escape from the endosome into the cytosol by a novel membrane translocation mechanism, which we have termed the “vesicle budding-and-collapse (VBC)” mechanism (Figure 3) [1-4]. We subsequently discovered that bacterial toxins, which are large, folded proteins, also escape the endosome by the VBC mechanism [5]. We have also shown that the cyclic CPPs can be used to effectively deliver all major drug modalities including small molecules, peptides, proteins, nucleic acids, and protein/nucleic acid complexes into the cytosol of mammalian cells. The cyclic CPPs have since become the scientific basis of a successful biotechnology company (Entrada Therapeutics).
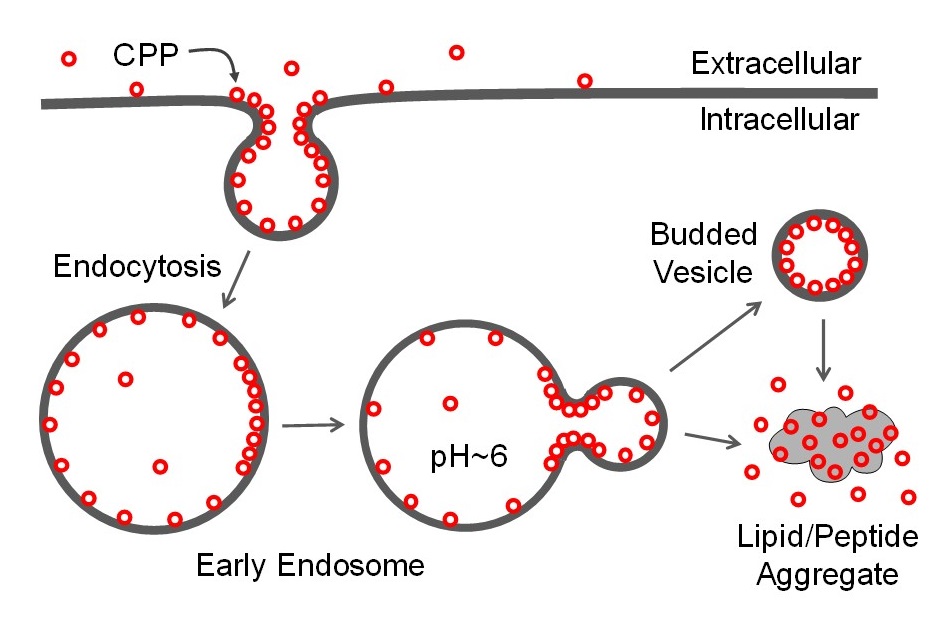
Figure 3. Mechanism of cellular entry and endosomal escape by CPPs. CPPs bind to plasma membrane phospholipids and/or other cell surface components and are brought into the early endosome by different endocytic and pinocytic mechanisms. As the endosomal pH decreases, the CPPs gain increased binding affinity to the limiting membrane, cluster into CPP-enriched lipid domains, which then bud off as small vesicles. Subsequent disintegration of the budded vesicle releases the luminal contents into the cytosol.
Current and future studies in this area include:
-
Mechanistic study of how other bacterial toxins and viruses enter the cell
-
Mechanistic study of how bacterial and eukaryotic proteins are secreted in their native states
-
Mechanism study of how cyclic CPPs and CPP-drug conjugates are eliminated from human cells
-
Designing genetically encodable cyclic CPPs for protein delivery and CPPs that selectively enter disease tissues/cells (e.g., cancer cells)
-
Engineer cell-permeable proteins as chemical probes and therapeutic agents [6]
Key Publications:
- Qian, Z., Martyna, A., Hard, R. L., Wang, J., Appiah-Kubi, G., Coss, C., Phelps, M. A., Rossman, J. S., and Pei, D. (2016) Discovery and Mechanism of Highly Efficient Cyclic Cell-Penetrating Peptides. Biochemistry 55, 2601-2612.
- Sahni, A., Qian, Z., and Pei, D. (2020) Cell-Penetrating Peptides Escape the Endosome by Inducing Vesicle Budding and Collapse. ACS Chem. Biol. 15, 2485-2492.
- Pei, D. (2022) How Do Biomolecules Cross the Cell Membrane? Chem. Res. 55, 309–318.
- Pei, D., and Dalbey, R. E. (2022) Membrane Translocation of Folded Proteins, Biol. Chem. 298, 102107.
- Sahni, A., and Pei, D. (2021) Bacterial Toxins Escape the Endosome by Inducing Vesicle Budding and Collapse. ACS Chem. Biol. 16, 2415–2422.
- Chen, K., and Pei, D. (2020) Engineering Cell-Permeable Proteins through Insertion of Cell-Penetrating Motifs into Surface Loops. ACS Chem. Biol. 15, 2568-2576.