Protein-Ligand Interactions
Protein-ligand interactions are critical to the function of many proteins in vivo. Characterization of the location of small molecule binding and ligand-induced conformational changes, along with quantifying the thermodynamics of such interactions, gives greater insight into the function of many diverse protein systems. Native mass spectrometry (nMS), coupled with collision-induced dissociation (CID), surface-induced dissociation (SID), electron-capture dissociation (ECD), and/or ion mobility (IM) offers a great way to explore protein-ligand interactions. We have published work showing the applicability of nMS for locating small molecule binding sites (figure 1), probing the impact of ligand binding on protein stability (figure 2), and quantifying protein-ligand interactions (figure 3).
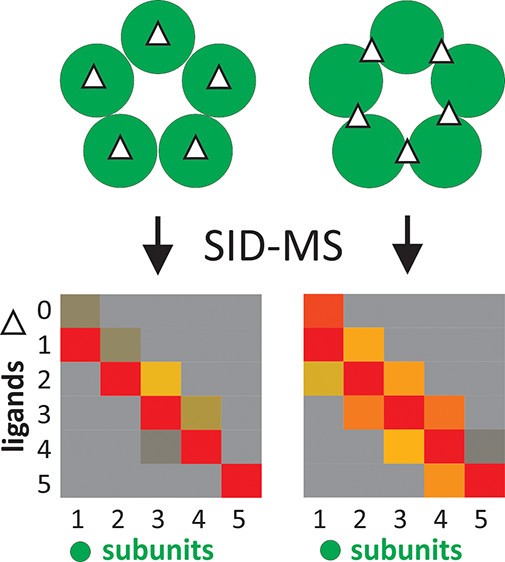
Figure 1. Ligand binding in CRP (left, ligand binding site within each subunit) and CTB (right, ligand binding site between each adjacent subunit). nMS and SID could be used in localizing sites of small ligand binding for multisubunit.1
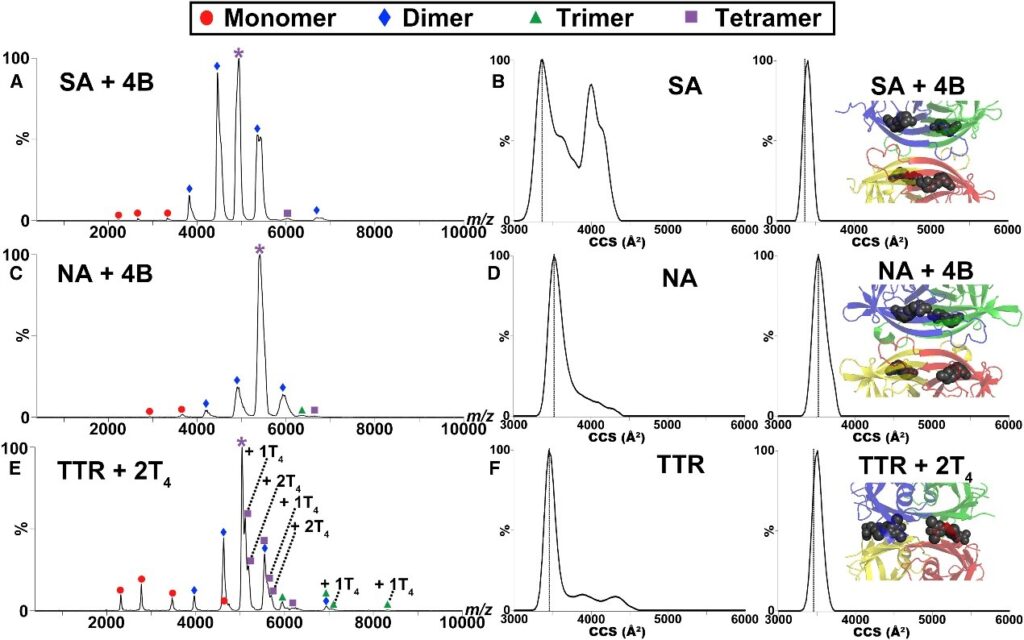
Figure 2. Ligand Binding Results in Stabilization of Native Homotetramer.2
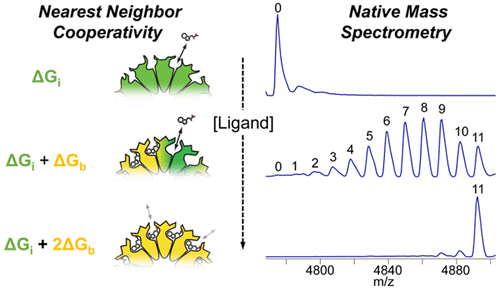
Figure 3. Quantification of the cooperativity of trp binding to the 11-mer ring-shaped protein TRAP.3
Reference:
- Busch, F.; VanAernum, Z. L.; Ju, Y.; Yan, J.; Gilbert, J. D.; Quintyn, R. S.; Bern, M.; Wysocki, V. H., Localization of protein complex bound ligands by surface-induced dissociation high-resolution mass spectrometry, Anal Chem. 2018, 90(21):12796-12801. DOI: 10.1021/acs.analchem.8b03263. [Request reprint © (pdf)]
- Quintyn, Royston S.; Yan, J.; Wysocki, Vicki H., Surface-Induced Dissociation of Homotetramers with D2 Symmetry Yields their Assembly Pathways and Characterizes the Effect of Ligand Binding. Chemistry & Biology 2015; 22 (5), 583-592. DOI: 10.1016/j.chembiol.2015.03.019. [Request reprint © (pdf)]
- Holmquist, M.; Ihms, E. C., Gollnick, P.; Wysocki, V. H.; and Foster, M. P.; Population distributions from native mass spectrometry titrations reveal nearest-neighbor cooperativity in the ring-shaped oligomeric protein TRAP. Biochemistry. 2020, 59, 27, 2518–252. DOI: 10.1021/acs.biochem.0c00352. [Request reprint © (pdf)]
