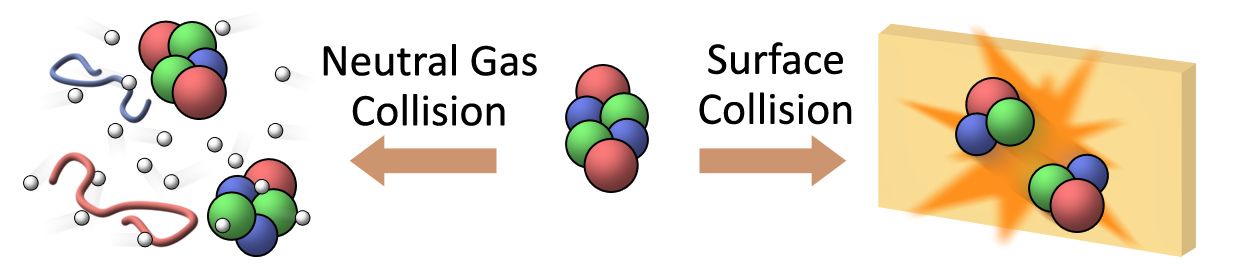
Subunit organization of protein complexes can be studied with tandem mass spectrometry by disruption of the quaternary structure in a controlled manner. The major challenge of using this method for structural analysis of non-covalent protein complexes is to overcome the undesired unfolding of subunits that occurs in the commonly used collision activation with gasous targets (Collision Induced Dissociation, CID), which results in loss of information on the original conformation. Our custom surface induced dissociation (SID) device has shown promising results of revealing sub-architecutre of protein complexes with minimal unfolding. The ultimate goal of this research is to develop a better mass spectrometric tool for improved structural characterization of large protein complexes.
Ongoing Projects in the group
- Surface induced dissociation coupled with ion mobility
- Protein-protein and Protein-ligand complexes
- Surface-induced dissociation of multimeric protein complexes (PDF)
- Chemical Crosslinking – Probing the interface of Proteins (PDF)
Previous Projects
Please click to download the PDF for detailed descriptions
- Surface-Induced dissociation in a quadrupole time-of-flight mass spectrometer
- Protein interactions probed by mass spectrometric techniques: hydrogen deuterium exchange, limited proteolysis and crosslinking
- Self-assembled monolayers (SAMs) as collision surfaces for ion activation
- Characterizing electron transfer properties of molecular wires through ion-surface collisions
- Surface-induced dissociation (SID) using derivatized silicon substrates
- Surface-induced dissociation kinetics
