- Han, D.; Spehar, J.M.; Richardson, D.S.; Leelananda, S.P.; Chakravarthy, P.; Grecco, S.; Reardon, J.; Stover, D.G.; Bennett, C.; Sizemore, G.M.; Li, Z.; Lindert, S.; Sizemore, S.T. (2024), The RAL Small G Proteins Are Clinically Relevant Targets in Triple Negative Breast Cancer. Cancers 16 (17), 3043. (Abstract | PubMed | PDF)
- Powell, J.W.; Mann, C.A.; Toth, P.D.; Nolan, S.; Steinert, A.; Ove, C.; Seffernick, J.T.; Wozniak, D.J.; Kebriaei, R.; Lindert, S.; Osheroff, N.; Yalowich, J.C.; Mitton-Fry, M.J. (2024), Development of Novel Bacterial Topoisomerase Inhibitors Assisted by Computational Screening. ACS Med Chem Lett 15 (8), 1287-1297. (Abstract | PubMed | PDF)
- Higgins, W.T.; Vibhute, S.; Bennett, C.; Lindert, S. (2024), Discovery of Nanomolar Inhibitors for Human Dihydroorotate Dehydrogenase Using Structure-Based Drug Discovery Methods. J Chem Inf Model 64 (2), 435-448. (Abstract | PubMed | PDF)
- Turzo, S.B.A.; Seffernick, J.T.; Lyskov, S.; Lindert, S. (2023), Predicting ion mobility collision cross sections using projection approximation with ROSIE-PARCS webserver. Brief Bioinform 24 (5), bbad308. (Abstract | PubMed | PDF)
- Kovvali, S.; Gao, Y.; Cool, A.M.; Lindert, S.; Wysocki, V.H.; Bell, C.E.; Gopalan, V. (2023), Insights Into The Catalytic Mechanism of A Bacterial Deglycase Essential For Utilization of Fructose-Lysine. Protein Sci 32 (7), e4695. (Abstract | PubMed | PDF)
- Cool, A.M.; Lindert, S. (2023), Umbrella Sampling Simulations of Cardiac Thin Filament Reveal Thermodynamic Consequences of Troponin I Inhibitory Peptide Mutations. J Chem Inf Model 63 (11), 3534-3543. (Abstract | PubMed | PDF)
- Hantz, E.R.; Tikunova, S.B.; Belevych, N.; Davis, J.P.; Reiser, P. J.; Lindert, S. (2023), Targeting Troponin C with Small Molecules Containing Diphenyl Moieties: Calcium Sensitivity Effects on Striated Muscles and Structure-Activity Relationship. J Chem Inf Model 63 (11), 3462-3473. (Abstract | PubMed | PDF)
- Drake, Z.C.; Seffernick, J.T.; Lindert, S. (2022), Protein complex prediction using Rosetta, AlphaFold, and mass spectrometry covalent labeling. Nat Commun 13 (1), 7846. (Abstract | PubMed | PDF)
- Hantz, E.R.; Lindert, S. (2022), Computational Exploration and Characterization of Potential Calcium Sensitizing Mutations in Cardiac Troponin C. J Chem Inf Model 62 (23), 6201-6208. (Abstract | PubMed | PDF)
- Hantz, E.R.; Lindert, S. (2022), Actives-Based Receptor Selection Strongly Increases the Success Rate in Structure-Based Drug Design and Leads to Identification of 22 Potent Cancer Inhibitors. J Chem Inf Model 62 (22), 5675-5687. (Abstract | PubMed | PDF)
- Turzo, S.B.A.; Hantz, E.R.; Lindert, S. (2022), Applications of machine learning in computer-aided drug discovery. QRB Discov 3, e14. (Abstract | PubMed | PDF)
- Thirugnanasambantham, P.; Kovvali, S.; Cool, A.M.; Gao, Y.; Sabag-Daigle, A.; Boulanger, E.F.; Mitton-Fry, M.J.; Capua, A.D.; Behrman, E.J.; Wysocki, V.H.; Lindert, S.; Ahmer, B.M.M.; Gopalan, V. (2022), Serendipitous Discovery of a Competitive Inhibitor of FraB, a Salmonella Deglycase and Drug Target. Pathogens 11 (10), 1102. (Abstract | PubMed | PDF)
- Cool, A.M.; Lindert, S. (2022), Umbrella Sampling Simulations Measure Switch Peptide Binding and Hydrophobic Patch Opening Free Energies in Cardiac Troponin. J Chem Inf Model 62 (22), 5666-5674. (Abstract | PubMed | PDF)
- He, J.; Turzo, S.B.A.; Seffernick, J.T.; Kim, S.S.; Lindert, S. (2022), Prediction of Intrinsic Disorder Using Rosetta ResidueDisorder and AlphaFold2. J Phys Chem B 126 (42), 8439-8446. (Abstract | PubMed | PDF)
- Turzo, S.B.A.; Seffernick, J.T.; Rolland, A.D.; Donor, M.T.; Heinze, S.; Prell, J.S.; Wysocki, V.H.; Lindert, S. (2022), Protein shape sampled by ion mobility mass spectrometry consistently improves protein structure prediction. Nat Commun 13 (1), 4377. (Abstract | PubMed | PDF)
- Rayani, K.; Hantz, E.R.; Haji-Ghassemi, O.; Li, A.Y.; Spuches, A.M.; Van Petegem, F.; Solaro, R.J.; Lindert, S.; Tibbits, G.F. (2022), The effect of Mg2+ on Ca2+ binding to cardiac troponin C in hypertrophic cardiomyopathy associated TNNC1 variants. FEBS J 289 (23), 7446-7465. (Abstract | PubMed | PDF)
- Seffernick, J.T.; Turzo, S.B.A.; Harvey, S.R.; Kim, Y.; Somogyi, Á.; Marciano, S.; Wysocki, V.H.; Lindert, S. (2022), Simulation of Energy-Resolved Mass Spectrometry Distributions from Surface-Induced Dissociation. Anal Chem 94 (29), 10506-10514. (Abstract | PubMed | PDF)
- Khaje, N.A.; Eletsky, A.; Biehn, S.E.; Mobley, C.K.; Rogals, M.J.; Kim, Y.; Mishra, S.K.; Doerksen, R.J.; Lindert, S.; Prestegard, J.H.; Sharp, J.S. (2022), Validated determination of NRG1 Ig-like domain structure by mass spectrometry coupled with computational modeling. Commun Biol 5 (1), 452. (Abstract | PubMed | PDF)
- Biehn, S.E.; Picarello, D.M., Pan, X.; Vachet, R.W.; Lindert, S. (2022), Accounting for Neighboring Residue Hydrophobicity in Diethylpyrocarbonate Labeling Mass Spectrometry Improves Rosetta Protein Structure Prediction. J Am Soc Mass Spectrom 33 (3), 584-591. (Abstract | PubMed | PDF)
- Nguyen, T.T.; Marzolf, D.R.; Seffernick, J.T.; Heinze, S.; Lindert, S. (2021), Protein structure prediction using residue-resolved protection factors from hydrogen-deuterium exchange NMR. Structure 30 (2), 313-320. (Abstract | PubMed | PDF)
- Biehn, S.E.; Lindert, S. (2021), Protein Structure Prediction with Mass Spectrometry Data. Annu Rev Phys Chem. 73, 1-19. (Abstract | PubMed | PDF)
- Lu, Y.; Vibhute, S.; Li, L.; Okumu, A.; Ratigan, S.C.; Nolan, S.; Papa, J.L.; Mann, C.A.; English, A.; Chen, A.; Seffernick, J.T.; Koci, B.; Duncan, L.R.; Roth, B.; Cummings, J.E.; Slayden, R.A.; Lindert, S.; McElroy, C.A.; Wozniak, D.J.; Yalowich, J.; Mitton-Fry, M.J. (2021), Optimization of TopoIV Potency, ADMET Properties, and hERG Inhibition of 5-Amino-1,3-dioxane-Linked Novel Bacterial Topoisomerase Inhibitors: Identification of a Lead with In Vivo Efficacy against MRSA. J Med Chem 64 (20), 15214-15249. (Abstract | PubMed | PDF)
- Cool, A.M.; Lindert, S. (2021), Computational Methods Elucidate Consequences of Mutations and Post-translational Modifications on Troponin I Effective Concentration to Troponin C. J Phys Chem B 125 (27), 7388-7396. (Abstract | PubMed | PDF)
- Hantz, E.R.; Lindert, S. (2021), Adaptive Steered Molecular Dynamics Study of Mutagenesis Effects on Calcium Affinity in the Regulatory Domain of Cardiac Troponin C. J Chem Inf Model 61 (6), 3052-3057. (Abstract | PubMed | PDF)
- Biehn, S.E.; Limpikirati, P.; Vachet, R.W.; Lindert, S. (2021), Utilization of Hydrophobic Microenvironment Sensitivity in Diethylpyrocarbonate Labeling for Protein Structure Prediction. Anal Chem 93 (23), 8188-8195. (Abstract | PubMed | PDF)
- Seffernick, J.T.; Canfield, S.M.; Harvey, S.R.; Wysocki, V.H.; Lindert, S. (2021), Prediction of Protein Complex Structure Using Surface-Induced Dissociation and Cryo-Electron Microscopy. Anal Chem 93 (21), 7596–7605. (Abstract | PubMed | PDF)
- Marzolf, D.R.; Seffernick, J.T.; Lindert, S. (2021), Protein Structure Prediction from NMR Hydrogen-Deuterium Exchange Data. J Chem Theory Comput 17 (4), 2619-2629. (Abstract | PubMed | PDF)
- Rayani, K.; Seffernick, J.T.; Li, A.Y.; Davis, J.P.; Spuches, A.M.; Van Petegem, F.; Solaro, R.J.; Lindert, S.; Tibbits, G.F. (2021), Binding of calcium and magnesium to human cardiac Troponin C. J Biol Chem 296, 100350. (Abstract | PubMed | PDF)
- Biehn, S.E.; Lindert, S. (2021), Accurate Protein Structure Prediction with Hydroxyl Radical Protein Footprinting Data. Nat Commun 12, 341. (Abstract | PubMed | PDF)
- Kim, S.S.; Alves, M.J.; Gygli, P.; Otero, J.; Lindert, S. (2021), Identification of novel cyclin A2 binding site and nanomolar inhibitors of cyclin A2-CDK2 complex. Curr Comput Aided Drug Des 17 (1), 57-68. (Abstract | PubMed | PDF)
- Seffernick, J.T.; Lindert, S. (2020), Hybrid methods for combined experimental and computational determination of protein structure. J Chem Phys 153 (24), 240901. (Abstract | PubMed | PDF)
- Lu, Y.; Papa, J.L.; Nolan, S.; English, A.; Seffernick, J.T.; Shkolnikov, N.; Powell, J.; Lindert, S.; Wozniak, D.J.; Yalowich, J.; Mitton-Fry, M.J. (2020), Dioxane-Linked Amide Derivatives as Novel Bacterial Topoisomerase Inhibitors against Gram-Positive Staphylococcus aureus. ACS Med Chem Lett 11 (12), 2446-2454. (Abstract | PubMed | PDF)
- Coldren, W.H.; Tikunova, S.B.; Davis, J.P.; Lindert, S. (2020), Discovery of Novel Small-Molecule Calcium Sensitizers for Cardiac Troponin C: A Combined Virtual and Experimental Screening Approach. J Chem Inf Model 60 (7), 3648-3661. (Abstract | PubMed | PDF)
- Leman, J.K.; Weitzner, B.D.; Lewis, S.M.; Adolf-Bryfogle, J.; Alam, N.; Alford, R.F.; Aprahamian, M.; Baker, D.; Barlow, K.A.; Barth, P.; Basanta, B.; Bender, B.J.; Blacklock, K.; Bonet, J.; Boyken, S.E.; Bradley, P.; Bystroff, C.; Conway, P.; Cooper, S.; Correia, B.E.; Coventry, B.; Das, R.; De Jong, R.M.; DiMaio, F.; Dsilva, L.; Dunbrack, R.; Ford, A.S.; Frenz, B.; Fu, D.Y.; Geniesse, C.; Goldschmidt, L.; Gowthaman, R.; Gray, J.J.; Gront, D.; Guffy, S.; Horowitz, S.; Huang, P.S.; Huber, T.; Jacobs, T.M.; Jeliazkov, J.R.; Johnson, D.K.; Kappel, K.; Karanicolas, J.; Khakzad, H.; Khar, K.R.; Khare, S.D.; Khatib, F.; Khramushin, A.; King, I.C.; Kleffner, R.; Koepnick, B.; Kortemme, T.; Kuenze, G.; Kuhlman, B.; Kuroda, D.; Labonte, J.W.; Lai, J.K.; Lapidoth, G.; Leaver-Fay, A.; Lindert, S.; Linsky, T.; London, N.; Lubin, J.H.; Lyskov, S.; Maguire, J.; Malmström, L.; Marcos, E.; Marcu, O.; Marze, N.A.; Meiler, J.; Moretti, R.; Mulligan, V.K.; Nerli, S.; Norn, C.; Ó’Conchúir, S.; Ollikainen, N.; Ovchinnikov, S.; Pacella, M.S.; Pan, X.; Park, H.; Pavlovicz, R.E.; Pethe, M.; Pierce, B.G.; Pilla, K.B.; Raveh, B.; Renfrew, P.D.; Burman, S.S.R.; Rubenstein, A.; Sauer, M.F.; Scheck, A.; Schief, W.; Schueler-Furman, O.; Sedan, Y.; Sevy, A.M.; Sgourakis, N.G.; Shi, L.; Siegel, J.B.; Silva, D.A.; Smith, S.; Song, Y.; Stein, A.; Szegedy, M.; Teets, F.D.; Thyme, S.B.; Wang, R.Y.; Watkins, A.; Zimmerman, L.; Bonneau, R. (2020), Macromolecular Modeling and Design in Rosetta: Recent Methods and Frameworks. Nat Methods 17 (7), 665-680. (Abstract | PubMed | PDF)
- Leelananda, S.P.; Lindert, S. (2020), Using NMR Chemical Shifts and Cryo-EM Density Restraints in Iterative Rosetta-MD Protein Structure Refinement. J Chem Inf Model 60 (5), 2522-2532. (Abstract | PubMed | PDF)
- Sengupta, A.; Wu, J.; Seffernick, J.T.; Sabag-Daigle, A.; Thomsen, N.; Chen, T.H.; Di Capua, A.; Bell, C.E.; Ahmer, B.M.M.; Lindert, S.; Wysocki, V.H.; Gopalan, V. (2019), Integrated use of biochemical, native mass spectrometry, computational and genome-editing methods to elucidate the mechanism of a Salmonella deglycase. J Mol Biol 431 (22), 4497-4513. (Abstract | PubMed | PDF)
- Seffernick, J.T.; Harvey, S.R.; Wysocki, V.H.; Lindert, S. (2019), Predicting Protein Complex Structure from Surface-Induced Dissociation Mass Spectrometry Data. ACS Cent Sci 5 (8), 1330-1341. (Abstract | PubMed | PDF)
- Bowman, J.D.; Lindert, S. (2019), Computational Studies of Cardiac and Skeletal Troponin. Front Mol Biosci 6:68. (Abstract | PubMed | PDF)
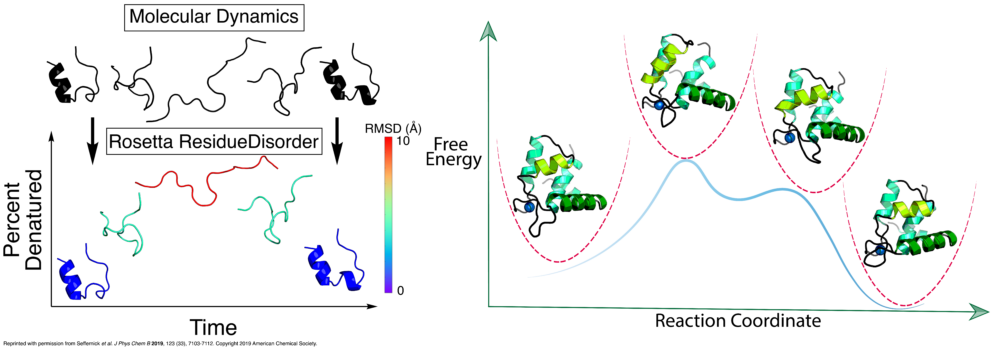
- Seffernick, J.T.; Ren, H.; Kim, S.S.; Lindert, S. (2019), Measuring Intrinsic Disorder and Tracking Conformational Transitions Using Rosetta ResidueDisorder. J Phys Chem B 123 (33), 7103-7112. (Abstract | PubMed | PDF)
- Bowman, J.D.; Coldren, W.H.; Lindert, S. (2019), Mechanism of Cardiac Troponin C Calcium Sensitivity Modulation by Small Molecules Illuminated by Umbrella Sampling Simulations. J Chem Inf Model 59 (6), 2964-2972. (Abstract | PubMed | PDF)
- Li, L.; Okumu, A.A.; Nolan, S.; English, A.; Vibhute, S.; Lu, Y.; Hervert-Thomas, K.; Seffernick, J.T.; Azap, L.; Cole, S.L.; Shinabarger, D.; Koeth, L.; Lindert, S.; Yalowich, J.; Wozniak, D.J.; Mitton-Fry, M.J. (2019), 1,3-Dioxane-linked Bacterial Topoisomerase Inhibitors with Enhanced Antibacterial Activity and Reduced hERG Inhibition. ACS Infect Dis 5 (7), 1115-1128. (Abstract | PubMed | PDF)
- Aprahamian, M.L.; Lindert, S. (2019), Utility of Covalent Labeling Mass Spectrometry Data in Protein Structure Prediction with Rosetta. J Chem Theory Comput 15 (5), 3410-3424. (Abstract | PubMed | PDF)
- Harvey, S.R.; Seffernick, J.T.; Quintyn, R.S.; Song, Y.; Ju, Y.; Yan, J.; Sahasrabuddhe, A.N.; Norris, A.; Zhou, M.; Behrman, E.J.; Lindert, S.; Wysocki, V.H. (2019), Relative interfacial cleavage energetics of protein complexes revealed by surface collisions. Proc Natl Acad Sci U S A 116 (17), 8143-8148. (Abstract | PubMed | PDF)
- Kim, S.S.; Aprahamian, M.L.; Lindert, S. (2019), Improving inverse docking target identification with Z-score selection. Chem Biol Drug Des 93 (6), 1105-1116. (Abstract | PubMed | PDF)
- Rhodes, C.A.; Dougherty, P.G.; Cooper, J.K.; Qian, Z.; Lindert, S.; Wang, Q.E.; Pei, D. (2018), Cell-Permeable Bicyclic Peptidyl Inhibitors against NEMO-IκB kinase Interaction Directly from a Combinatorial Library. J Am Chem Soc 140 (38), 12102–12110. (Abstract | PubMed | PDF)
- Bowman, J.D.; Lindert, S. (2018), Molecular Dynamics and Umbrella Sampling Simulations Elucidate Differences in Troponin C Isoform and Mutant Hydrophobic Patch Exposure. J Phys Chem B 122 (32), 7874–7883. (Abstract | PubMed | PDF)
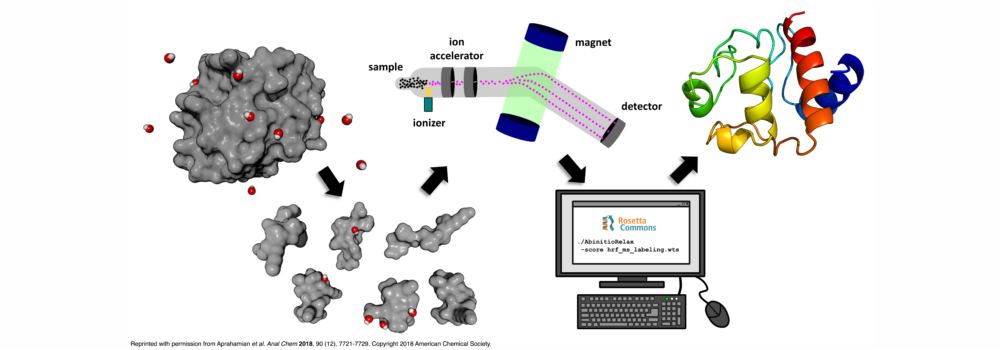
- Aprahamian, M.L.; Chea, E.E.; Jones, L.M.; Lindert, S. (2018), Rosetta Protein Structure Prediction from Hydroxyl Radical Protein Footprinting Mass Spectrometry Data. Anal Chem 90 (12), 7721–7729. (Abstract | PubMed | PDF)
- Kim, S.S.; Seffernick, J.T.; Lindert, S. (2018), Accurately Predicting Disordered Regions of Proteins Using Rosetta ResidueDisorder Application. J Phys Chem B 122 (14), 3920-30. (Abstract | PubMed | PDF)
- Aprahamian, M.L.; Tikunova, S.B.; Price, M.V.; Cuesta, A.F.; Davis, J.P.; Lindert, S. (2017), Successful Identification of Cardiac Troponin Calcium Sensitizers Using a Combination of Virtual Screening and ROC Analysis of Known Troponin C Binders. J Chem Inf Model 57 (12), 3056-3069. (Abstract | PubMed | PDF)
- Leelananda, S.P.; Lindert, S. (2017), Iterative Molecular Dynamics-Rosetta Membrane Protein Structure Refinement Guided by Cryo-EM Densities. J Chem Theory Comput 13 (10), 5131-5145. (Abstract | PubMed | PDF)
- Schwebach, C.L.; Agrawal, R.; Lindert, S.; Kudryashova, E.; Kudryashov, D.S. (2017), The Roles of Actin-Binding Domains 1 and 2 in the Calcium-Dependent Regulation of Actin Filament Bundling by Human Plastins. J Mol Biol 429 (16), 2490–2508. (Abstract | PubMed | PDF)
- Leelananda, S.P.; Lindert, S. (2016), Computational methods in drug discovery. Beilstein J Org Chem 12, 2694–2718. (Abstract | PubMed | PDF)
- Cai, F.; Li, M. X.; Pineda-Sanabria, S.E.; Gelozia, S.; Lindert, S.; West, F.; Sykes, B.D.; Hwang, P.M. (2016), Structures reveal details of small molecule binding to cardiac troponin. J Mol Cell Cardiol 101, 134-44. (Abstract | PubMed | PDF)
- Dewan, S.; McCabe, K. J.; Regnier, M.; McCulloch, A. D.; Lindert, S. (2016), Molecular Effects of cTnC DCM Mutations on Calcium Sensitivity and Myofilament Activation – An Integrated Multi-Scale Modeling Study. J Phys Chem B 120 (33), 8264-75. (Abstract | PubMed | PDF)
- Cheng, Y.; Lindert, S.; Oxenford, L.; Tu, A.Y.; McCulloch, A. D.; Regnier, M. (2016), The Effects of Cardiac Troponin I Mutation P83S on Contractile Properties and the Modulation by PKA-Mediated Phosphorylation. J Phys Chem B 120 (33), 8238-53. (Abstract | PubMed | PDF)
- Feng, X.; Zhu, W.; Schurig-Briccio, L.A.; Lindert, S.; Shoen, C.; Hitchings, R.; Li, J.; Wang, Y.; Baig, N.; Zhou, T.; Kim, B.K.; Crick, D.C.; Cynamon , M.; McCammon, J. A.; Gennis , R.B.; Oldfield, E. (2015), Antiinfectives targeting enzymes and the proton motive force. Proc Natl Acad Sci U S A 112 (51), E7073-E7082. (Abstract | PubMed | PDF)
- Cheng, Y.; Rao, V. S.; Tu, A.Y.; Lindert, S.; Wang, D.; Oxenford, L.; McCulloch, A. D.; McCammon, J. A.; Regnier, M. (2015), Troponin I Mutations R146G and R21C Alter Cardiac Troponin Function, Contractile Properties, and Modulation by Protein Kinase A (PKA)-mediated Phosphorylation. J Biol Chem 290 (46), 27749-27766. (Abstract | PubMed | PDF)
- Lindert, S.; McCammon, J. A. (2015), Improved cryoEM-Guided Iterative Molecular Dynamics–Rosetta Protein Structure Refinement Protocol for High Precision Protein Structure Prediction. J Chem Theory Comput 11 (3), 1337–1346. (Abstract | PubMed | PDF)
- Lindert, S.; Cheng, Y.; Kekenes-Huskey, P. M.; Regnier, M.; McCammon, J. A. (2015), Effects of HCM cTnI Mutation R145G on Troponin Structure and Modulation by PKA Phosphorylation elucidated by Molecular Dynamics Simulations. Biophys J 108 (2), 395–407. (Abstract | PubMed | PDF)
- Lindert, S.; Tallorin, L. C.; Nguyen, Q. G.; Burkart, M.D.; McCammon, J. A. (2015), In silico Screening for Plasmodium falciparum Enoyl-ACP Reductase inhibitors. J Comput Aided Mol Des 29 (1), 79-87. (Abstract | PubMed | PDF)
- Lindert, S.; Li, M. X.; Sykes, B.; McCammon, J.A. (2015), Computer-aided drug discovery approach finds calcium sensitizer of cardiac troponin. Chem Biol Drug Des 85 (2), 99-106. (Abstract | PubMed | PDF)
Article featured as Editor’s Choice - Zinsser, V. L.; Lindert, S.; Banford, S.; Hoey, E. M.; Trudgett, A.; Timson, D. J. (2015), UDP-galactose 4′-epimerase from the liver fluke, Fasciola hepatica: biochemical characterisation of the enzyme and identification of inhibitors. Parasitology 142 (3), 463-472. (Abstract | PubMed | PDF)
- Kim, M. O.; Feng, X.; Feixas, F.; Wei, Z.; Lindert, S.; Bogue, S.; Sinko, W.; de Oliveira, C.; Rao, G.; Oldfield, E.; McCammon, J. A. (2015), A Molecular Dynamics Investigation of Mycobacterium tuberculosis Prenyl Synthases: Conformational Flexibility, and Implications for Computer-Aided Drug Discovery. Chem Biol Drug Des 85(6):756-69. (Abstract | PubMed | PDF)
- Cheng, Y.; Lindert, S.; Kekenes-Huskey, P. M.; Rao, V. S.; Solaro, R.J.; Rosevear, P.R.; Amaro, R.; McCulloch, A. D.; McCammon, J. A.; Regnier, M. (2014), Computational Studies of S23D/S24D Troponin I Mutation on Cardiac Troponin Structural Dynamics. Biophys J 107 (7), 1675-1685. (Abstract| PubMed | PDF)
- Rao, V. S.; Cheng, Y.; Lindert, S.; Wang, D.; Oxenford, L.; McCulloch, A. D.; McCammon, J. A.; Regnier, M. (2014), PKA phosphorylation of cardiac troponin I modulates activation and relaxation kinetics of ventricular myofibrils. Biophys J 107 (5), 1196-1204. (Abstract | PubMed | PDF)
- Liu, Y. L.*; Lindert, S.*; Zhu, W.; Wang, K.; McCammon, J. A.; Oldfield, E. (2014), Taxodione and arenarone inhibit farnesyl diphosphate synthase by binding to the isopentenyl diphosphate site. Proc Natl Acad Sci U S A 111 (25), E2530-E2539. (Abstract | PubMed | PDF)
- Goetz, A.; Bucher, D.; Lindert, S.; McCammon, J. A. (2014), Dipeptide aggregation in aqueous solution from fixed point-charge force fields. J Chem Theory Comput 10 (4), 1631–1637 (Abstract | PubMed | PDF)
- Lindert, S.*; Maslennikov I.*; Chiu E.; Pierce, L.C.; McCammon, J. A.; Choe, S. (2014), Drug screening strategy for human membrane proteins: From NMR protein backbone structure to in silica- and NMR-screened hits. Biochem Biophys Res Commun 445 (4), 724-733 (Abstract | PubMed | PDF)
- Feixas, F.; Lindert, S.; Sinko, W.; McCammon, J. A. (2014), Exploring the Role of Receptor Flexibility in Structure-Based Drug Discovery. Biophysical Chemistry 186C, 31-45 (Abstract | PubMed | PDF)
- Lindert, S.; Bucher, D.; Eastman, P.; Pande, V.; McCammon, J. A. (2013), Accelerated Molecular Dynamics Simulations with the AMOEBA Polarizable Force Field on Graphics Processing Units. J Chem Theory Comput 9 (11), 4684–4691 (Abstract | PubMed | PDF)
- Lindert, S.; Meiler, J.; McCammon, J. A. (2013), Iterative Molecular Dynamics-Rosetta Protein Structure Refinement Protocol to Improve Model Quality. J Chem Theory Comput 9 (8), 3843-3847 (Abstract | PubMed | PDF)
- Durrant, J. D.; Lindert, S.; McCammon, J. A. (2013), AutoGrow 3.0: An improved algorithm for chemically tractable, semi-automated protein inhibitor design. J Mol Graph Model 44C, 104-112 (Abstract | PubMed | PDF)
- Timson, D.; Lindert, S. (2013), Comparison of Dynamics of Wildtype and V94M Human UDP-Galactose 4-Epimerase – A computational perspective on severe Epimerase-deficiency Galactosemia. Gene 526 (2), 318-24 (Abstract | PubMed | PDF)
- Lindert, S.; Zhu, W.; Liu, Y. L.; Pang, R.; Oldfield, E.; McCammon, J.A. (2013), Farnesyl diphosphate synthase inhibitors from in silico screening. Chem Biol Drug Des 81 (6), 742-8. (Abstract | PubMed | PDF)
- Zhu, W.; Zhang, Y.; Sinko, W.; Hensler, M. E.; Olson, J.; Molohon, K. J.; Lindert, S.; Cao, R.; Li, K.; Wang, K.; Wang, Y.; Liu, Y. L.; Sankovsky, A.; de Oliveira, C. A.; Mitchell, D. A.; Nizet, V.; McCammon, J. A.; Oldfield, E. (2013), Antibacterial drug leads targeting isoprenoid biosynthesis. Proc Natl Acad Sci U S A 110 (1), 123-128. (Abstract | PubMed | PDF)
- Sinko, W.; Lindert, S.; McCammon, J. A. (2013), Accounting for Receptor Flexibility and Enhanced Sampling Methods in Computer-Aided Drug Design. Chem Biol Drug Des 81 (1), 41-49. (Abstract | PubMed | PDF)
- Lindert, S.; Stewart, P. L.; Meiler, J. (2013), Computational determination of the orientation of a heat repeat-like domain of DNA-PKcs. Comput Biol Chem 42, 1-4. (Abstract | PubMed | PDF)
- Lindert, S.; Kekenes-Huskey, P. M.; McCammon, J. A. (2012). Long-timescale molecular dynamics simulations elucidate the dynamics and kinetics of exposure of the hydrophobic patch in troponin C. Biophys J 103 (8), 1784-1789. (Abstract | PubMed | PDF)
- Lindert, S. and McCammon, J.A. (2012). Dynamics of Plasmodium falciparum enoyl-ACP reductase and implications on drug discovery. Protein Sci 21(11): 1734-1745. (Abstract | PubMed | PDF)
- Kekenes-Huskey, P. M., Lindert, S. and McCammon, J.A. (2012). Molecular basis of calcium-sensitizing and desensitizing mutations of the human cardiac troponin C regulatory domain: a multi-scale simulation study. PLoS Comput Biol 8(11). (Abstract | PubMed | PDF)
- Lindert, S., Kekenes-Huskey, P.M., Huber, G., Pierce, L., and McCammon, J.A. (2012). Dynamics and calcium association to the N-terminal regulatory domain of human cardiac troponin C: a multiscale computational study. J Phys Chem B 116, 8449-8459. (Abstract | PubMed | PDF)
- Lindert, S.*, Durrant, J.*, McCammon, J. A. (2012). LigMerge: A Fast Algorithm to Generate Models of Novel Potential Ligands from Sets of Known Binders. Chem Biol Drug Des 80, 358-365. (Abstract | PubMed | PDF)
- Lindert, S., Alexander, N., Wotzel, N., Karakas, M., Stewart, P.L., and Meiler, J. (2012). EM-Fold: De Novo Atomic-Detail Protein Structure Determination from Medium-Resolution Density Maps. Structure 20, 464-478. (Abstract | PubMed | PDF)
Article featured in PSI Nature Structural Biology KB - Lindert, S., Hofmann, T., Wotzel, N., Karakas, M., Stewart, P.L., and Meiler, J. (2012). Ab initio protein modeling into CryoEM density maps using EM-Fold. Biopolymers 97, 669-677. (Abstract | PubMed | PDF)
- Woetzel, N., Lindert, S., Stewart, P.L., and Meiler, J. (2011). BCL::EM-Fit: rigid body fitting of atomic structures into density maps using geometric hashing and real space refinement. Journal of Structural Biology 175, 264-276. (Abstract | PubMed | PDF)
- Smith JG, Silvestry M, Lindert, S., Lu W, Nemerow GR, et al. (2010) Insight into the Mechanisms of Adenovirus Capsid Disassembly from Studies of Defensin Neutralization. PLoS Pathog 6(6). (Abstract | PubMed | PDF)
- Lindert, S., Stewart, P.L., and Meiler, J. (2009). Hybrid approaches: applying computational methods in cryo-electron microscopy. Curr Opin Struct Biol 19, 218-225. (Abstract | PubMed | PDF)
- Lindert, S., Staritzbichler, R., Wotzel, N., Karakas, M., Stewart, P.L., and Meiler, J. (2009). EM-fold: De novo folding of alpha-helical proteins guided by intermediate-resolution electron microscopy density maps. Structure 17, 990-1003. (Abstract | PubMed | PDF)
- Lindert, S., Silvestry, M., Mullen, T.M., Nemerow, G.R., and Stewart, P.L. (2009). Cryo-electron microscopy structure of an adenovirus-integrin complex indicates conformational changes in both penton base and integrin. J Virol 83, 11491-11501. (Abstract | PubMed | PDF)
- Silvestry, M., Lindert, S., Smith, J.G., Maier, O., Wiethoff, C.M., Nemerow, G.R., and Stewart, P.L. (2009). Cryo-electron microscopy structure of adenovirus type 2 temperature-sensitive mutant 1 reveals insight into the cell entry defect. J Virol 83, 7375-7383. (Abstract | PubMed | PDF)
* signifies equal contribution